Camera Trap Images
Camera Trap Images#
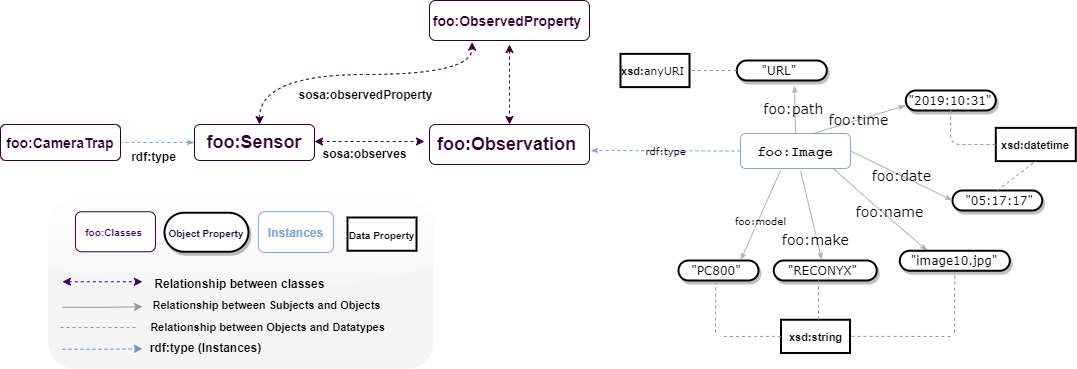
A dataset containing one thousand images of Elephas maxims was modelled. Before transforming them into RDF graphs, image metadata was extracted and saved as CSV files. The RDF dataset has a unique path that points to the location of the image on a protected cloud server. The figure below depicts the dataset entities and the output of the semantic modelling. Data are accessable on Github (https://github.com/Naeima/Forest-Observatory-Ontology/tree/main/Camera Trap images metadata).
!pip install rdflib
from google.colab import files
import pandas as pd
from rdflib import Graph, Namespace, URIRef, Literal
from rdflib.namespace import RDF, XSD
import csv
# Prompt for file upload
uploaded = files.upload()
# Read the uploaded CSV file into a pandas DataFrame
for filename in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=filename, length=len(uploaded[filename])))
df = pd.read_csv(filename)
print(df.head())
# Define namespaces
FOO = Namespace("https://w3id.org/def/foo#")
SOSA = Namespace("http://www.w3.org/ns/sosa/")
GEO = Namespace("http://www.w3.org/2003/01/geo/wgs84_pos#")
# Create a new graph
g = Graph()
# Bind namespaces
g.bind("foo", FOO)
g.bind("sosa", SOSA)
g.bind("geo", GEO)
# Path to your CSV file
csv_file_path = filename # Adjusted to dynamically handle the uploaded CSV file
# Read and process data from the image CSV
with open(csv_file_path, newline='') as csvfile:
reader = csv.DictReader(csvfile, delimiter=',')
# Print headers for debugging
print("Headers:", reader.fieldnames)
for row in reader:
image_id = row.get('Image_ID', '').strip() # Assuming there's an 'Image_ID' field
timestamp = row.get('Timestamp', '').strip() # Assuming there's a 'Timestamp' field
camera_id = row.get('Camera_ID', '').strip() # Assuming there's a 'Camera_ID' field
location = row.get('Location', '').strip() # Assuming there's a 'Location' field
species = row.get('Species', '').strip() # Assuming there's a 'Species' field
# Create an observation URI for the image
observation = URIRef(f"https://w3id.org/def/foo#image_{image_id}")
g.add((observation, RDF.type, FOO.imageObservation))
g.add((observation, FOO.imageID, Literal(image_id, datatype=XSD.string)))
g.add((observation, FOO.timestamp, Literal(timestamp, datatype=XSD.dateTime)))
g.add((observation, FOO.cameraID, Literal(camera_id, datatype=XSD.string)))
g.add((observation, FOO.location, Literal(location, datatype=XSD.string)))
if species:
g.add((observation, FOO.observes, Literal(species, datatype=XSD.string)))
# Serialize the graph to a file
output_file = "image_metadata_knowledge_graph.ttl"
g.serialize(destination=output_file, format="turtle")
print(f"Knowledge graph has been serialized to {output_file}")
# Download the RDF Turtle file
from google.colab import files
files.download(output_file)
```python
# adding serialized data to stardog
# conn_details = {
# 'endpoint': 'http://localhost:5820',
# 'username': 'admin',
# 'password': 'admin'
# }
# with stardog.Admin(**conn_details) as admin:
# images = admin.new_database('images')
# conn = stardog.Connection('images', **conn_details)
# conn.begin()
# conn.add(
# stardog.content.File('images.rdf', stardog.content_types.TURTLE),
# )
# conn.commit()
