FOO Publication
Contents
FOO Publication#
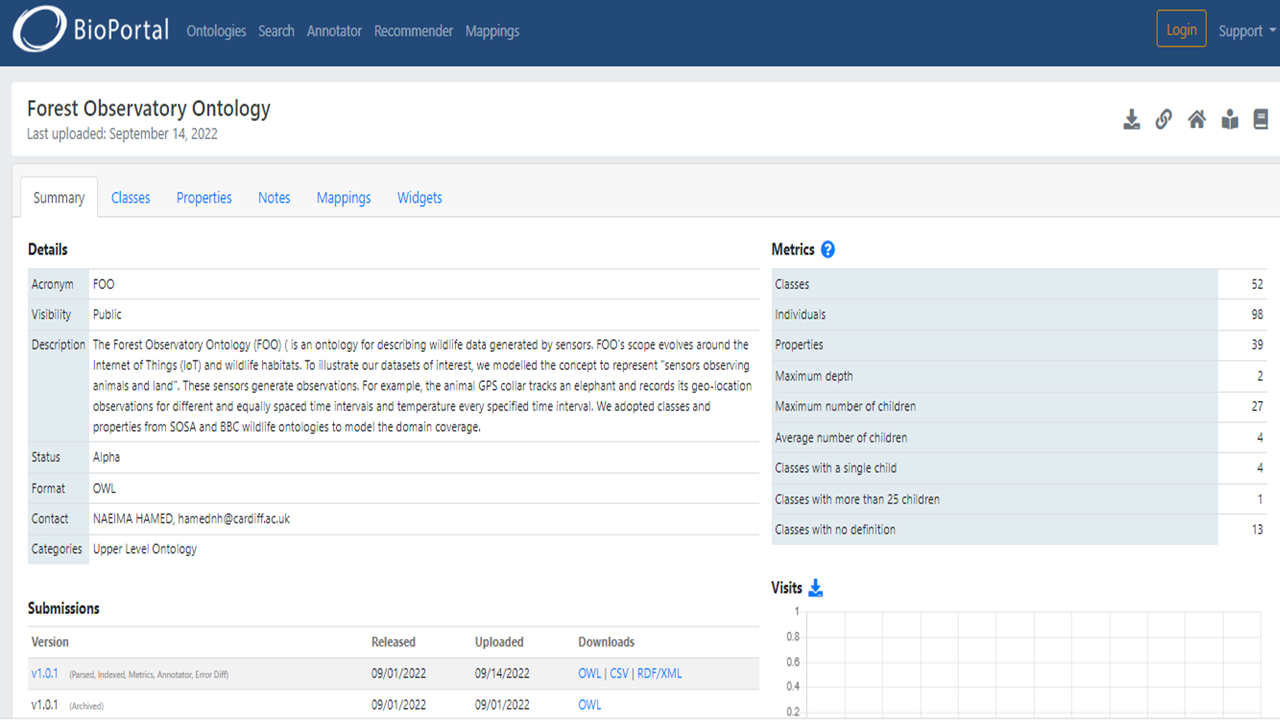
Ontologies are typically created in editors and then exported in formats (e.g., Turtle, RDF/XML, JSON-LD) that are challenging for others to use. Researchers sometimes refer to articles or technical reports explaining ontology to solve this challenge. However, papers usually emphasise the scientific contribution rather than defining each concept in the ontology. Documenting the ontology entities might be a solution. For this reason, semantic web communities have developed tools to assist with ontology documentation. These tools extract annotation properties from OWL ontologies and generate HTML documentation for classes, properties, and instances. We chose to use the WIZARD for DOCumenting Ontology (WIDOCO) https://dgarijo.github.io/Widoco/doc/tutorial/ to document FOO as it builds on Live OWL Documentation Environment (LODE) used in the seven-star linked data model platform. WIDOCO software produces HTML pages containing human and machine-interpretable visualisation and Oops! (https://oops.linkeddata.es/) evaluation. Then we made FOO, and its documentation available in Github, not only to maintain it but for also sharing it with the research community, enabling them to contribute to its enhancement and interoperability (i.e., use it in software applications) To make FOO findable on the web in different interoperable formats after publishing, we followed W3C best practices and deposited it in the BioPortal repository. FOO is now active online under the creative commons Attribution 4.0 International (CC 4.0).
Github#
Bioportal#